

AI Data Scientist
Your AI-powered data science assistant
Sourcetable's AI Data Scientist enables you to perform advanced statistical analysis, machine learning, and predictive modeling on your spreadsheet data. Get instant insights with Python's data science libraries - no coding required.
Try for Free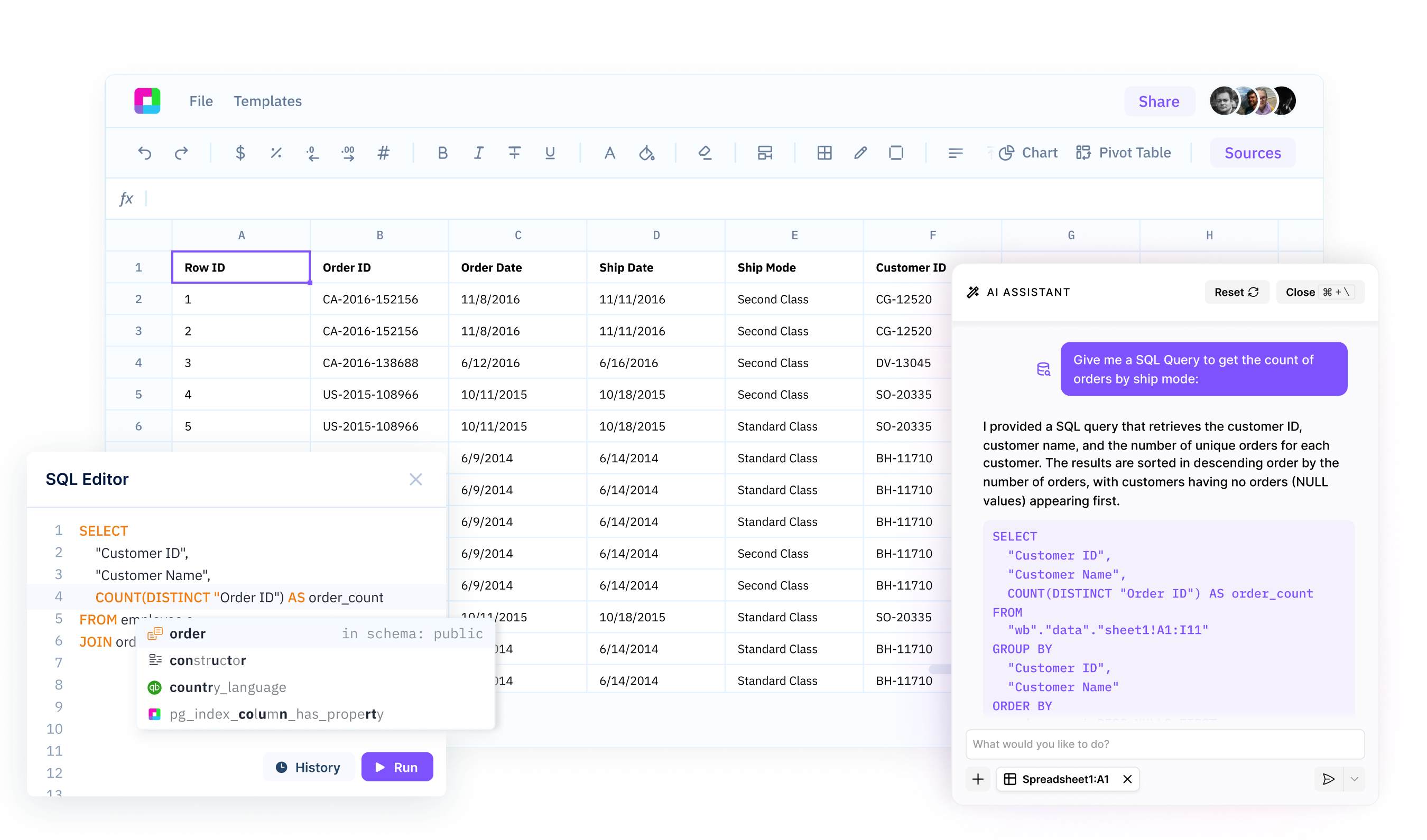
Machine learning without the complexity
Build, train, and evaluate machine learning models using scikit-learn, XGBoost, and statsmodels. Ask in plain English and get production-ready ML code instantly.
Advanced statistical analysis made simple
Perform hypothesis testing, regression analysis, time series forecasting, and multivariate statistics. Get insights from your data without a statistics PhD.
Feature engineering on autopilot
Automatically engineer features, handle missing data, scale variables, and encode categories. Let AI handle the data preprocessing pipeline.
Build Models and Find Patterns with AI
Run machine learning, statistical analysis, and advanced analytics using natural language. AI handles the complexity - focus on insights, not code.
Professional data science capabilities in a spreadsheet
Natural Language to Data Science
Describe your analysis goals in plain English and Sourcetable generates the corresponding Python code using pandas, scikit-learn, scipy, and statsmodels. Build predictive models, run A/B tests, or perform clustering analysis without writing code.
Try for free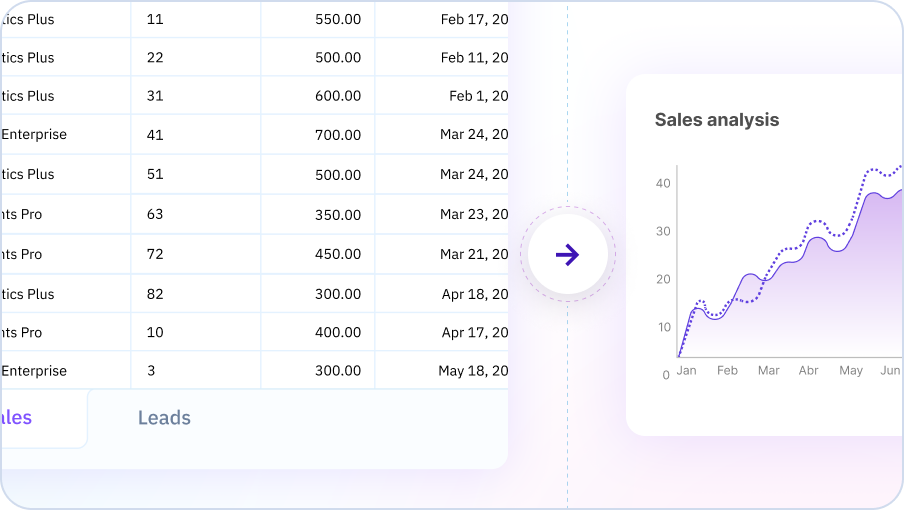
Intelligent Analysis Recommendations
Sourcetable's AI analyzes your data structure and suggests appropriate statistical tests, ML algorithms, and visualization techniques. Get expert guidance on which methods work best for your specific dataset and research questions.
Try for free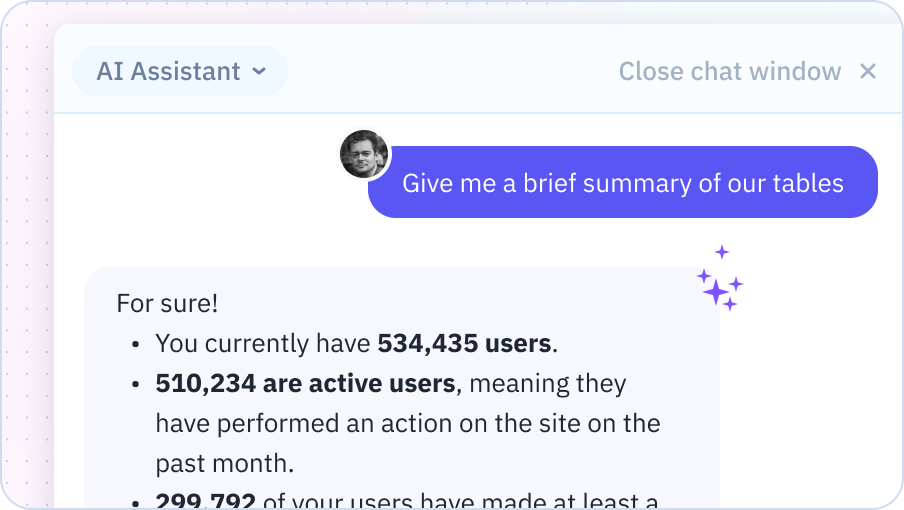
End-to-End ML Pipeline
From data preprocessing to model deployment, execute complete machine learning workflows in your spreadsheet. Train models, tune hyperparameters, evaluate performance, and generate predictions - all with AI assistance.
Try for free
Data science workflows powered by AI
Build predictive models, perform statistical analysis, and engineer features - all without leaving your spreadsheet.
Democratize data science across your organization
Machine learning for everyone
Enable business analysts and domain experts to build predictive models without knowing Python or ML theory. Empower your team to make data-driven predictions and recommendations.
Production-ready code
Get clean, well-structured Python code that follows data science best practices. Export your analysis to Jupyter notebooks or integrate with existing ML pipelines.
Complete ML toolkit
Access the full ecosystem of Python data science libraries: scikit-learn for ML, statsmodels for statistics, scipy for scientific computing, and more. All without installation or setup.
Reproducible research
Every analysis is documented and reproducible. Share your work with colleagues who can understand, verify, and build upon your data science findings.
Team Purple
See what people say about Sourcetable

Chris Aubuchon
@ChrisAubuchon
Spreadsheets are still the best interface for so many real world projects, it's time for @SourcetableApp to give them a reboot

Micah Alpern
@malpern
Love seeing innovation in this space after so many decades with very little.
Frequently Asked Questions
What data science tasks can the AI perform?
Sourcetable's AI Data Scientist performs statistical analysis, machine learning model building, predictive modeling, feature engineering, data preprocessing, model evaluation, clustering, classification, regression, time series forecasting, and hypothesis testing - all through natural language commands.
Do I need Python or data science skills to use this?
No coding or data science expertise required. Describe your data science goal in plain English (like 'predict customer churn' or 'segment customers') and Sourcetable's AI handles the technical implementation using Python libraries like scikit-learn, pandas, and statsmodels.
What machine learning models can the AI build?
Sourcetable's AI can build classification models (Random Forest, Logistic Regression, Decision Trees), regression models (Linear, Ridge, Lasso), clustering algorithms (K-means, DBSCAN), and ensemble methods. It automatically selects appropriate algorithms based on your data and problem type.
Can the AI explain its data science approach?
Yes, Sourcetable's AI explains every step including data preprocessing, feature selection, algorithm choice, hyperparameter tuning, and model evaluation. It shows you the code it wrote and interprets model results in business terms.
How does the AI validate machine learning models?
Sourcetable's AI automatically performs train-test splits, cross-validation, and calculates performance metrics (accuracy, precision, recall, F1, RMSE, R²). It ensures models generalize well and aren't overfitted.
Can the AI handle feature engineering?
Yes, Sourcetable's AI performs feature scaling, encoding categorical variables, handling missing values, creating interaction features, and polynomial features. It prepares your data for optimal model performance automatically.
Frequently Asked Questions
How does AI Data Scientist work?
Sourcetable's AI Data Scientist uses advanced AI to analyze your data and provide intelligent insights. Simply upload your data or connect your database, then interact with Sourcetable's AI using natural language. Sourcetable's AI understands your requests and performs complex analysis without requiring technical expertise or coding knowledge.
Do I need technical skills to use AI Data Scientist?
No technical skills are required. AI Data Scientist is designed for users of all skill levels. You can interact with your data using plain English commands, and Sourcetable's AI handles all the complex technical work. Whether you're a beginner or an expert, Sourcetable makes data analysis accessible and intuitive.
What types of data can I analyze with AI Data Scientist?
AI Data Scientist works with a wide variety of data sources including CSV files, Excel spreadsheets (.xlsx, .xls), databases (PostgreSQL, MySQL, SQL Server), and many popular business applications. You can upload files directly or connect to live data sources through Sourcetable's connector system.
Can I export my results from AI Data Scientist?
Yes, you can export your analysis results in multiple formats including Excel (.xlsx), CSV, PDF, and other common formats. All charts, tables, and insights generated by AI Data Scientist can be downloaded and shared with your team or stakeholders.
Is my data secure when using AI Data Scientist?
Yes, Sourcetable takes data security seriously. Your data is protected by enterprise-grade security standards including SOC 2 Type 2, HIPAA, and PCI compliance. All data is encrypted at rest and in transit. Sourcetable's AI models are not trained on your data, ensuring your information remains private and confidential.
How is AI Data Scientist different from traditional spreadsheet tools?
Unlike traditional spreadsheet tools that require manual formulas and complex functions, AI Data Scientist uses AI to understand your questions and automatically perform analysis. Instead of spending hours learning functions or writing code, you can simply describe what you need in plain English and get instant results. This makes advanced data analysis accessible to everyone, not just spreadsheet experts.



